Genome Editing Technology Overview
To study the function of a gene, scientists usually manipulate its expression level by RNAi, which provides an easy and fast way to down regulate gene expression. However, some uncertainties always entwine RNAi technology. First, RNAi may only partially interfere the expression level and therefore some effect is hidden. Second, the off targets of RNAi is always a concern.
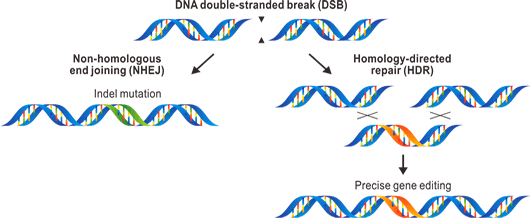 Genome editing provides the most reliable and versatile way to manipulate gene expression. What makes genome editing possible are nonhomologous end jointing (NHEJ) repairing and homology-directed repair (HDR) induced by DNA double-strand break (DSB). Genome editing provides the most reliable and versatile way to manipulate gene expression. What makes genome editing possible are nonhomologous end jointing (NHEJ) repairing and homology-directed repair (HDR) induced by DNA double-strand break (DSB).
-
NHEJ disrupts gene expression by creating some bases insertion or deletion, called indel mutations. One DSB repaired by NHEJ induce frame shift, and two DSB repaired by NHEJ produce a fragment deletion. For screening convenience, we involved any visible marker carried by transposable elements in two DSB CRISPR.
-
HDR functions as a versatile knock-in tool to bring a fragment in the gene-of-interested to create premature stop, to delete large gene region and replace it with an attP-landing site for further modifications, to bring in RMCE cassette for further gene exchange or reporter (such as Gals4) exchange, to replace some amino acids, and to fuse a fluorescence protein for trace. We are able to accept customized designs for your research needs.These methods allow scientists to manipulate the gene-of-interested as what they want at genome level once they can create DNA double-strand break specifically on the gene-of-interested. ZFN (Zinc-finger nuclease), TALEN (Transcription activator-like effector nuclease) and CRISPR (clustered regularly interspaced short palindromic repeat) are exactly what scientists need.
-
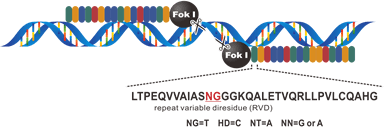 TALEN shows higher specificity and activity on double-strand breaks than ZFN, and started a genome editing new era in 2012. The target site of TALEN is composed of over 20 base nucleotides, usually 30~40 bases. Each nucleotide is recognized by a tandem array of 33~35 amino acid repeats. Specifically, the specificity of each repeat domain is determined by the 12th and 13th amino acids, which are called RVD (repeat variable di-residues). However, TALEN cloning is a time-, effort- and money-consuming step since scientists need to put over 20 35-amino-acid-repeats on the same construct. TALEN shows higher specificity and activity on double-strand breaks than ZFN, and started a genome editing new era in 2012. The target site of TALEN is composed of over 20 base nucleotides, usually 30~40 bases. Each nucleotide is recognized by a tandem array of 33~35 amino acid repeats. Specifically, the specificity of each repeat domain is determined by the 12th and 13th amino acids, which are called RVD (repeat variable di-residues). However, TALEN cloning is a time-, effort- and money-consuming step since scientists need to put over 20 35-amino-acid-repeats on the same construct.
-
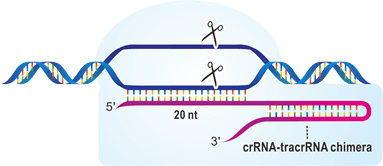 CRISPR, chosen as "Method of the Year" by both Nature and Science in 2013, is an immune system of bacteria. CRISPR make genome double stand break easy and specific. CRISPR function as a RNA-guided engineered nuclease, and is composed of crRNA, tracRNA and Cas9 nuclease. crRNA is processed from pre-crRNA under the contribution of tracRNA, and the 20-base sequence on crRNA is responsible for the specificity of target site. DualRNA-Cas9 then form an active DNA endonuclease to recognize 20 bases + NGG sequence on genome and undergo endonuclease activity. This makes CRISPR an easier, faster, cheaper way than TALEN to edit genome. Its efficiency is also superior to ZFN. Replacing TALEN, CRISPR is the hottest genome editing tool. CRISPR, chosen as "Method of the Year" by both Nature and Science in 2013, is an immune system of bacteria. CRISPR make genome double stand break easy and specific. CRISPR function as a RNA-guided engineered nuclease, and is composed of crRNA, tracRNA and Cas9 nuclease. crRNA is processed from pre-crRNA under the contribution of tracRNA, and the 20-base sequence on crRNA is responsible for the specificity of target site. DualRNA-Cas9 then form an active DNA endonuclease to recognize 20 bases + NGG sequence on genome and undergo endonuclease activity. This makes CRISPR an easier, faster, cheaper way than TALEN to edit genome. Its efficiency is also superior to ZFN. Replacing TALEN, CRISPR is the hottest genome editing tool.

Want to Have Your Specific CRISPR Fly?
Simply choose WellGenetics as your research partner!
We will propose the best strategy and do the rest of steps for you - from Design to Fly!
Check Our Services Now
|


 Genome editing provides the most reliable and versatile way to manipulate gene expression. What makes genome editing possible are nonhomologous end jointing (NHEJ) repairing and homology-directed repair (HDR) induced by DNA double-strand break (DSB).
Genome editing provides the most reliable and versatile way to manipulate gene expression. What makes genome editing possible are nonhomologous end jointing (NHEJ) repairing and homology-directed repair (HDR) induced by DNA double-strand break (DSB). TALEN shows higher specificity and activity on double-strand breaks than ZFN, and started a genome editing new era in 2012. The target site of TALEN is composed of over 20 base nucleotides, usually 30~40 bases. Each nucleotide is recognized by a tandem array of 33~35 amino acid repeats. Specifically, the specificity of each repeat domain is determined by the 12th and 13th amino acids, which are called RVD (repeat variable di-residues). However, TALEN cloning is a time-, effort- and money-consuming step since scientists need to put over 20 35-amino-acid-repeats on the same construct.
TALEN shows higher specificity and activity on double-strand breaks than ZFN, and started a genome editing new era in 2012. The target site of TALEN is composed of over 20 base nucleotides, usually 30~40 bases. Each nucleotide is recognized by a tandem array of 33~35 amino acid repeats. Specifically, the specificity of each repeat domain is determined by the 12th and 13th amino acids, which are called RVD (repeat variable di-residues). However, TALEN cloning is a time-, effort- and money-consuming step since scientists need to put over 20 35-amino-acid-repeats on the same construct.  CRISPR, chosen as "Method of the Year" by both Nature and Science in 2013, is an immune system of bacteria. CRISPR make genome double stand break easy and specific. CRISPR function as a RNA-guided engineered nuclease, and is composed of crRNA, tracRNA and Cas9 nuclease. crRNA is processed from pre-crRNA under the contribution of tracRNA, and the 20-base sequence on crRNA is responsible for the specificity of target site. DualRNA-Cas9 then form an active DNA endonuclease to recognize 20 bases + NGG sequence on genome and undergo endonuclease activity. This makes CRISPR an easier, faster, cheaper way than TALEN to edit genome. Its efficiency is also superior to ZFN. Replacing TALEN, CRISPR is the hottest genome editing tool.
CRISPR, chosen as "Method of the Year" by both Nature and Science in 2013, is an immune system of bacteria. CRISPR make genome double stand break easy and specific. CRISPR function as a RNA-guided engineered nuclease, and is composed of crRNA, tracRNA and Cas9 nuclease. crRNA is processed from pre-crRNA under the contribution of tracRNA, and the 20-base sequence on crRNA is responsible for the specificity of target site. DualRNA-Cas9 then form an active DNA endonuclease to recognize 20 bases + NGG sequence on genome and undergo endonuclease activity. This makes CRISPR an easier, faster, cheaper way than TALEN to edit genome. Its efficiency is also superior to ZFN. Replacing TALEN, CRISPR is the hottest genome editing tool.